Lung cancer is the leading cause of cancer-related deaths worldwide, with approximately 2.2 million new cases and 1.8 million deaths reported in 2020.[1] It represents a group of heterogeneous entities in terms of histology and molecular profile.[2] Primary lung cancers are classified into two main histological types: Non-small cell lung carcinoma (NSCLC) and small cell lung carcinoma (SCLC), representing 85% and 15% of all cases, respectively. NSCLC can be further divided into subtypes, including lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and large-cell carcinoma.[2] Low survival rates for both NSCLC and SCLC have been linked to late diagnosis and subpar response to conventional chemotherapy and radiation. This emphasizes how crucial it is to identify novel biomarkers and possible therapeutic targets to enhance the prognosis of lung cancer patients. Despite all the NSCLCs have molecularly characterized pre-invasive lesions, they are often asymptomatic and undetectable by the current noninvasive methods. Lots of researches are in progress to identify lung cancer early detection methods. Unfortunately, most of them are imaging technology-based ones and low-dose computed tomography has been identified as a sensitive technique for the early detection of stage 0 lung cancers, and this has been suggested as a screening method. Sputum is a desirable prospective source of lung cancer biomarkers due to its noninvasive nature and propensity to represent the site of injury.[3] Various research teams have examined sputum cytology as a potential technique for early lung cancer detection.[4] However, none of these studies were found fruitful as sputum cytology suffers a lack of sensitivity to be used for early detection. The reason for the lower sensitivity was the lack of adequate laboratory techniques to fish out the whole cell content without any morphological distortion. Furthermore, it was challenging to distinguish between reactive benign cells and malignant cells due to the morphological changes brought about by laboratory processing techniques, as well as the inherent reactive and degenerative changes caused by the exfoliated cells clogged on mucus. There were no other methods available to enhance the morphological assessment. Hence, the cytologist used to report the same as to contain atypical cells. An average of 20–30% of sputum samples are being reported to have such atypical cells without a definite diagnosis in the pathology division of our hospital.[5] However, now, the liquid-based cytology (LBC) methods offer better technology to extract the whole cell content of sputum samples without much distortion. Furthermore, there are ancillary techniques to supplement the conventional method. We recently developed a modified sputum processing method that preserves cells for additional molecular analyses and enables the characterization of specific proteins to complement conventional morphological evaluation.[5]
Initiating DNA replication and cell proliferation requires the presence of minichromosome maintenance proteins (MCMs), a group of proteins involved in minichromosome maintenance. They are referred to as the DNA replication-licensing factors by joining the origin recognition complex with additional licensing factors to form the pre-replicative complex, which regulates DNA replication and only permits it to take place once every cell cycle.[6] In addition, they play a role in the cohesion, condensation, transcription, and recombination of DNA molecules and are necessary for replication elongation. A trimeric core complex made up of the MCM4, MCM6, and MCM7 subunits is coupled with a peripheral dimer of the MCM3 and MCM5 to produce a hexameric MCM complex.[7] Throughout the cell cycle, all MCM proteins are invariably present in the nucleus. Because MCM proteins are essential for initiating DNA replication in proliferating cells but are absent in quiescent cells, they serve as reliable markers of cell proliferation.[8] In a number of cancers, such as lung, ovarian, colon, urothelial, and oral cancers, the expression level of MCMs has been linked to important clinicopathological parameters and demonstrated considerable diagnostic and prognostic value.[9-11] In addition, MCMs may serve as indicators for recurrent and precancerous illnesses, according to a number of lines of research, and abnormal expressions of MCMs have been linked to the onset and development of several cancers.[9,12] Particularly in humans and other eukaryotes, MCM2 and MCM7 are engaged in a variety of biological processes.[13] MCM2 is highly expressed during the proliferation of premalignant lung cells and is considered a potential diagnostic marker for the early detection of malignant pulmonary lesions, whereas MCM7 expression in bronchial brushings of NSCLC patients is associated with prognosis.[14] However, the application of MCM7 protein evaluation in sputum cytology still remains unfathomed.
The current work sought to characterize malignant cells in sputum using MCM2 and MCM7. Through the identification of these proteins, the research aimed to improve traditional sputum cytology. Furthermore, the expression of these proteins was investigated in relation to various clinicopathological characteristics.
MATERIAL AND METHODS SubjectsA cohort of 97 patients who were referred from the Medical College Hospital and Sanatorium for Chest Disease in 2019 and 2020 served as the study’s subjects. The individuals ranged in age from 39 to 79 years old and included both genders. The selection of subjects was based on:
Inclusion criteriaAdults aged 18 years and above
Positive sputum cytology for malignancy confirmed by biopsy
Negative sputum cytology findings
Complete clinical diagnoses, including, at a minimum, negative chest X-rays
Patients capable of producing adequate sputum samples for cytological analysis
Patients who provided informed consent for participation.
Exclusion criteriaPatients who have received prior treatment for lung cancer
Patients with other significant pulmonary diseases, such as tuberculosis or pulmonary fibrosis
Sputum samples that are insufficient in quantity or quality (fewer than five pulmonary macrophages)
Patients with a history of other malignancies.
19 LUAD, 23 LUSC, 10 NSCLC, 24 atypical, and 21 with negative for malignant cells (NMCs) are among the lesions that have been histologically shown to be malignant. The selection of these cases was predicated on the sufficiency of their sputum samples and positive clinical follow-up information. Of them, 32 cases belonged to stage III, 22 to stage IV, and 22 to stage II. Clinical complaints and additional clinico-pathological data were collected from patient records and documented on a proforma. Informed consent was obtained from each participant following approval of the study by the institutional review board and human ethics committee (HEC No. 13/019). The study was conducted in accordance with the Declaration of Helsinki.
Sample collection and processingEarly morning sputum for 3 consecutive days was obtained in the sample bottle provided to the patients. Post-bronchoscopic samples were collected from patients selected for bronchoscopy. Ninety-seven samples of sputum have been selected for the study. Cytologically positive sputum samples with biopsy confirmation of malignancy were taken as positive, and subjects with negative cytology and complete clinical diagnosis, which includes, as a minimum, negative chest X-rays were taken as negative. All three samples of each patient preserved at 4°C were pooled, homogenized, and treated with BD Cytorich® red Preservative (#491336, Becton, Dickinson and Company, USA). After vortexing the samples with twice the volume of red preservative, they were left for half an hour. The combined sample was transferred to a 50 mL centrifuge tube, vortexed again, and centrifuged for 5 min at 600 g. The resulting cell pellet was re-suspended in buffer solution, vortexed, and centrifuged again for 10 min at 800 g. This pellet was then used for cell block preparation.[5] Samples for immunocytochemistry have been selected based on the cytopathological analysis for sample adequacy and the presence of representative cells.
ImmunocytochemistryImmunocytochemistry was carried out in 5 micron sections from cell blocks using MACH4 Universal horseradish peroxidase (HRP) polymer detection system (#BRR4012, Biocare Medical, Pacheco, CA 94553, USA). The sections were deparaffinized with three changes of xylene (CAS no: 1330-20-7, Sigma -Aldrich Chemical Pvt. Limited Industrial Area, Anekal Taluka Plot No 12, Bangalore, India), each for 10 min, followed by rehydration through decreasing concentrations of alcohol and a final rinse in water, each step lasting 5 min. Cell permeability was enhanced by treating with 4 mM sodium deoxycholate (#RM131, HiMedia, HiMedia Laboratories Pvt. Limited, Maharashtra, India) for 15 min. Antigen retrieval was carried out using the microwave method with sodium citrate buffer (pH 6.0) at 700 W for 15 min. After washing the slides for 5 min in deionized water, the endogenous peroxidase activity was blocked using Peroxidazed 1 (#PX968H, Biocare Medical, Pacheco, CA 94553, USA) for 5 min. Slides were washed twice in TBST (TRIS-buffered saline-Tween20 [#MB067, HiMedia, HiMedia Laboratories Pvt. Limited, Maharashtra, India]). The samples were then incubated with a background punisher (#BRR974H, Biocare Medical, Pacheco, CA 94553, USA) for protein block for 5 min and then washed twice in TBST for 5 min each. Sections were treated with primary antibody overnight at 4°C. Mouse monoclonal primary antibodies MCM2 (# sc-373702, Santha Cruz Biotechnology, Texas 75220, USA) and MCM7 (# sc-65469, Santha Cruz Biotechnology, Texas 75220, USA) were used. Antibodies were used in a dilution of 1:25. After washing in TBST, the sections were incubated with MACH4 mouse probe (#BRR534DH, Biocare medical, Pacheco, CA 94553, USA) for 30 min at room temperature and rinsed in TBST twice for 5 min each. The sections were incubated for 30 min at room temperature with MACH4 HRP polymer (#BRR534BH, Biocare Medical, Pacheco, CA 94553, USA) and rinsed twice TBST for 5 min each. Peroxidase activity was developed using 3, 3’ Diaminobenzidine (#BRR900AC, Biocare Medical, Pacheco, CA 94553, USA). Slides were rinsed in deionized water and then counterstained with Harris Hematoxylin (GRM9946, Biocare Medical, Pacheco, CA 94553, USA) followed by bluing in running tap water, dehydrated in ascending grades of alcohol for 5 min each, then cleared in three changes of xylene and mounted in dibutylphthalate polystyrene xylene (#100579, Sigma-Aldrich, Merck, Mumbai, India).
The immunocytochemically stained slides were analyzed using a Leica light microscope. Positive control slides were first screened to recognize the pattern of expression. Each of the immunostained slides was initially screened at ×10 magnification followed by ×20 magnification for grading the different intensities of expression pattern. The intensity of expression patterns was scored as mild, moderate, and intense on a zero to 3 + scale. H-scores, ranging from 0 to 300, were calculated by multiplying the staining intensity (0–3) by the percentage of positive cells (0–100%). An H-score of 30 or higher was considered positive. Two of the investigators carried out the immunoscoring separately. Rescoring was done on samples if there was any disagreement regarding immunoscores. Images were captured with a camera system attached to the microscope, utilizing the Leica application software (Leica Application LA Suite V3 [LASV3.8], Leica Microsystems, Danaher, Germany).
Statistical analysisThe statistical analysis for the study was conducted using the Statistical Package for the Social Sciences software (version 28.0, IBM Corporation; Armonk, NY, USA). Descriptive statistics were employed to summarize the demographic and clinical characteristics of the study participants, with frequencies and percentages applied to the categorical variables. When the theoretical frequency T ≥ 5 and sample size N ≥ 40, Chi-square test was used; when 1 ≤ T < 5 and N ≥ 40, continuity-corrected Chi-square was used for the test; and when T = 0, Fisher Freeman Halton exact test was used. To evaluate the diagnostic performance of each marker, with the cytology report as the gold standard, sensitivity and specificity were calculated, along with positive predictive value (PPV) and negative predictive value. To assess the diagnostic efficacy of the sputum markers, receiver operating characteristic (ROC) curve was generated and the area under the curve (AUC) was calculated. An AUC value closer to 1 indicates a better diagnostic performance, with AUC >0.9 indicating high accuracy. A p-value threshold of <0.05 was set for all statistical tests. Results with P-values below this threshold were considered statistically significant.
RESULTS Baseline characteristics and expression patterns of MCM proteinsThe study included 97 subjects, comprising 81.4% males (79 out of 97) and 18.6% females (18 out of 97). Among these subjects, 64.9% (63 out of 97) were smokers. The clinicopathological features of the 97 study subjects provided important insights into the association between MCM2 and MCM7 protein expression and different lung cancer subtypes. MCM7-negative and MCM7-positive groups did not exhibit a significant difference with respect to age (P = 0.540) and sex (P = 0.246) in the different subgroups. Similarly, MCM2 and MCM7 expression did not show significant variation between smokers and non-smokers (P = 0.785 and P = 0.799, respectively). Of the 34 non-smokers, 22 were positive for MCM2, while 24 were positive for MCM7. Similarly, among smokers (63 subjects), 39 were MCM2 positive, and 46 were MCM7 positive, suggesting that smoking status does not have a strong association with MCM protein expression. Among the 19 patients with LUAD, 16 were positive for MCM2, and all were positive for MCM7, showing a strong association (P < 0.001) for both markers. All 23 LUSC cases were positive for both MCM2 and MCM7, further reinforcing the diagnostic relevance of these markers in this subtype (P < 0.001 for both). Among the 10 NSCLC cases, all were positive for both MCM2 and MCM7 (P < 0.001), indicating that MCM proteins are strongly expressed in this subtype as well. Out of 24 atypical cases, 12 were positive for MCM2, and 18 were positive for MCM7, with a significant correlation for MCM7 (P < 0.001). The total sample size for the clinical stage analysis is 76 because the 21 samples without malignant cells were not assigned a clinical stage. Among the 22 patients in Stage II, 17 were positive for MCM2 and MCM7, though only MCM7 expression reached statistical significance (P = 0.014). This indicates that MCM7 might be more sensitive for detecting cancer at earlier stages. Of the 32 patients in Stage III, 24 were MCM2 positive and 31 were MCM7 positive. While MCM2 did not show significant correlation (P = 0.361), MCM7 was strongly associated with tumor stage, suggesting that MCM7 expression increases with disease progression. In Stage IV patients (22 total), 20 were positive for MCM2, and all were positive for MCM7, showing a stronger correlation between MCM7 and advanced-stage disease (P = 0.014). These findings suggest that MCM7 may be a more versatile marker, providing insights into both the type and progression of the tumor as shown in Table 1.
Table 1: Association of MCM2 and MCM7 expression with diverse clinicopathological characteristics in lung cancer.
Clinicopathological features n(97) MCM2 protein expression status P-value MCM7 protein expression status P-value Negative Positive Test Statistic Negative Positive Test Statistic Mean age (SD) 60.69 (9.19) 61.49 (7.582) −0.462 0.645 60.37 (9.96) 61.51 (7.43) −0.615 0.540 Gender Male 79 27 52 1.573 0.210 20 59 1.344 0.246 Female 18 9 9 7 11 Smoking status Non-Smoker 34 12 22 0.074 0.785 10 24 0.065 0.799 Smoker 63 24 39 17 46 Cytology LUAD 19 3 16 68.056 <0.001 0 19 75.333 <0.001 LUSC 23 0 23 0 23 NSCLC 10 0 10 0 10 Atypical 24 12 12 6 18 NMC 21 21 0 21 0 Clinical stage II 22 5 17 2.279 0.361 5 17 7.367 0.014 III 32 8 24 1 31 IV 22 2 20 0 22Due to the limited sample size within each tumor subtype, characterizing and comparing them individually with clinicopathological features would likely yield statistically insignificant results. Hence, such comparisons were avoided. Instead, a cumulative analysis was performed across all subtypes, examining protein expressions to ensure more robust and meaningful insights.
Intensity of MCM expression in different tumor typesThe expressions of MCM2 and MCM7 were confined exclusively to the nuclei, with no background staining observed. A positive MCM2 marker was detected in 62.9% of the subjects (61 out of 97), while overexpression of MCM7 was observed in 72.2% of the subjects (70 out of 97). The intensity of expression varied among different tumor types. Notably, LUAD samples exhibited the highest expression of MCM7, with a mean H-score of 184.3 [Table 2 and Supplementary Figure 1].
Table 2: Mean H score/SD of markers for different lung lesions in sputum samples.
Diagnosis Number of specimens MCM 2 mean H score (SD) MCM 7 mean H score (SD) LUAD 19 123.9 (70.3) 184.3 (66.3) LUSC 23 93.1 (53.5) 155.2 (68.9) NSCLC 10 77 (37.6) 113 (39.2) Atypical cells 24 22.5 (10.7) 53.5 (30.9) NMC 21 0 (0) 0 (0)The patterns of MCM2 and MCM7 expression showed varying degrees of intensity, including mild, moderate, and dense expressions in tumor cell nuclei. With a few exceptions, where weak localized expression was detected in a minimal set of cells, neither marker was expressed in samples that were negative for malignancy [Figure 1]. Hematoxylin and eosin (H&E) staining demonstrated the cellular morphology of LUAD, LUSC, NSCLC, atypical cells, and non-malignant cells ([Figure 1a, d, g, j, m], respectively). Moderate nuclear expression of MCM2 [Figure 1b] and dense nuclear expression of MCM7 [Figure 1c] were observed in adenocarcinoma cells. In squamous cell carcinoma cells, moderate nuclear expression of MCM2 and MCM7 was noted [Figure 1e and f]. Mild nuclear expression of both markers was seen in NSCLC cells [Figure 1h and i], while atypical cells exhibited weak or mild nuclear expression of MCM2 and MCM7 [Figure 1k and l], though this was limited to a small number of cells. In contrast, non-malignant samples were negative for both MCM2 and MCM7 [Figure 1n and o].
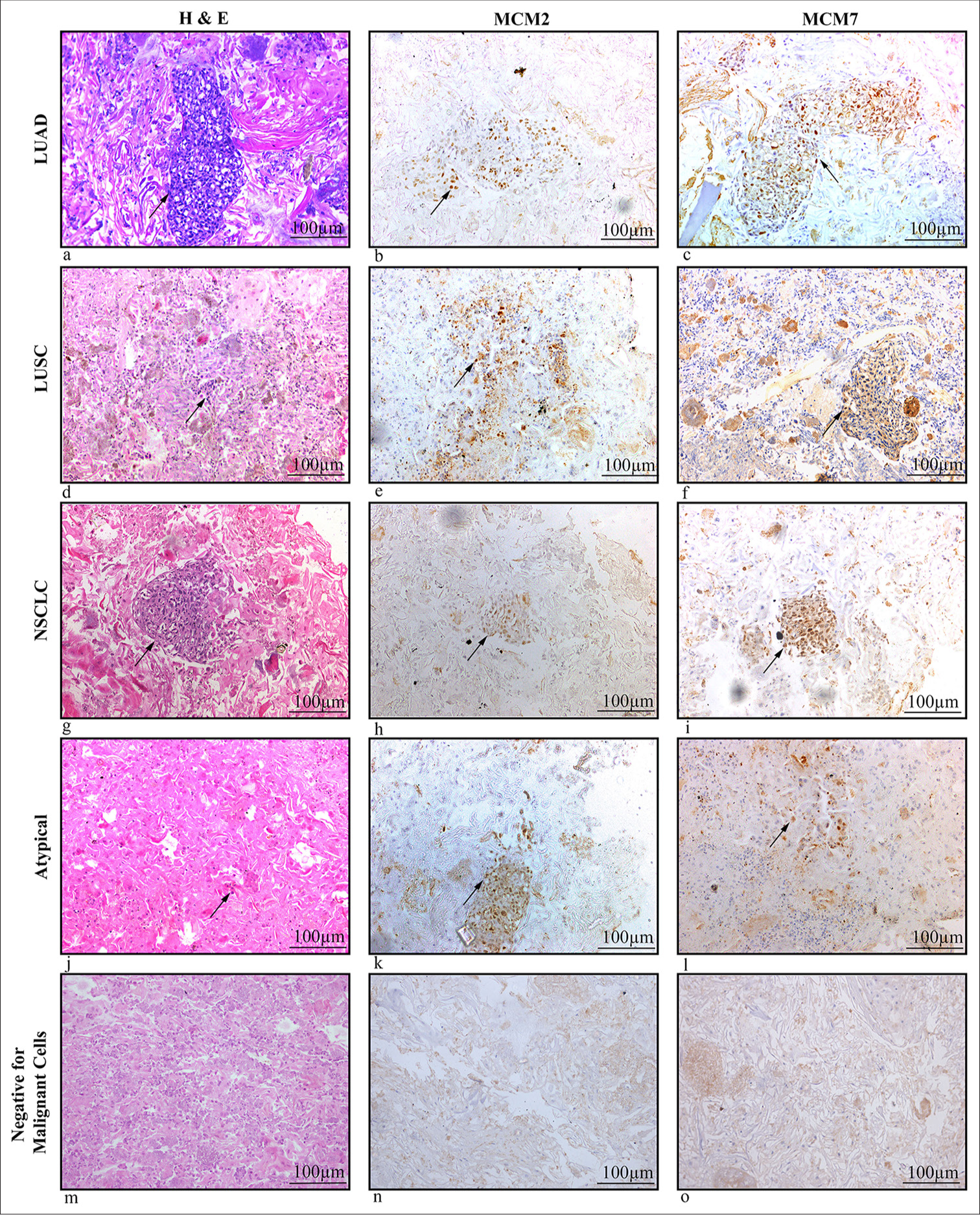
Export to PPT
Diagnostic performance of MCM2 and MCM7The diagnostic performance of MCM2 and MCM7 proteins for detecting malignancy was thoroughly evaluated. A total of 97 study subjects were tested, with 76 having malignancy and 21 being non-malignant. MCM2 demonstrated a sensitivity of 80.26% (61 true positives and 15 false negatives) and PPV of 100%, with a specificity of 100% (21 true negatives and 0 false positives), indicating that it is highly effective in correctly identifying malignant cases when present. On the other hand, MCM7 exhibited a higher sensitivity of 92.11% (70 true positives and 6 false negatives), also with a PPV of 100% and a specificity of 100% (21 true negatives and 0 false positives), suggesting that it is even more reliable in identifying malignant cells. To further assess the diagnostic accuracy, an ROC curve was constructed (figure not included as Table 3 is explanatory). The AUC for MCM7 was found to be 0.961, which is higher compared to MCM2, indicating superior overall diagnostic performance. This high AUC value for MCM7 underscores its robustness as a biomarker for malignancy in lung cancer diagnostics [Table 3].
Table 3: Diagnostic test result of MCM proteins based on H score.
Markers Sensitivity (%) Specificity (%) PPV (%) NPV (%) Accuracy AUC MCM 2 80.26 100 100 58.33 84.54 0.901 MCM 7 92.11 100 100 77.78 98.81 0.961 DISCUSSIONReplicative immortality and sustained proliferative signaling are the well-established hallmarks of cancer.[15,16] DNA replication is a key factor in cell proliferation, which occurs more frequently in malignant cells compared to healthy ones. Cancer cells have aberrant expression of several proteins involved in DNA replication. One example is the replication factor MCM7, which forms the pre-replication complex during the progression of the cell cycle and attaches to the double-stranded DNA at the replication origins in the late G1 phase.[17] It is highly expressed in various malignancies and may serve as a potential marker for cell proliferation.[18,19] Further, MCM7 has been demonstrated as a proliferation marker in several cancers, including lymphoma, ovarian cancer, prostate cancer, lung cancer, etc., equivalent to the established proliferation markers like Kiel-67 (Ki-67) and proliferating cell nuclear antigen.[10]
MCM proteins are more sensitive markers of proliferation than Ki-67 because MCM proteins can identify a broader range of proliferative cells, including those that may not be detected by Ki-67, especially in the early G1 phase.[20] MCM proteins have been studied in various types of lung cancers, including NSCLC and SCLC. Their expression is often higher in more aggressive forms. Ki-67 is also used across different lung cancer types, but its expression can differ significantly among diverse tumor types and even within distinct areas of the same tumor.[21] A meta-analysis by Martin et al. examined the prognostic value of Ki-67 expression in lung cancer, providing a comprehensive overview of its role in predicting patient outcomes.[22] Another study by Stoeber et al. investigated the diagnostic potential of MCM proteins in cancer, suggesting broader applications beyond lung cancer.[23] The use of MCM proteins in clinical practice is still evolving, but they offer potential as diagnostic and prognostic markers, possibly complementing or even surpassing Ki-67 in certain cases.
MCM7, unlike Ki-67, is expressed throughout the entire cell cycle (except G0), making it a more sensitive marker for detecting cells that are actively proliferating. This is particularly important in lung cancer, where the early detection of proliferative cells can significantly influence prognosis and treatment strategies. Williams G. H. and Stoeber K. emphasized the broader sensitivity of MCM proteins, including MCM7, in identifying proliferative cells across different phases of the cell cycle, which could improve the accuracy of diagnosis of lung cancer. In addition to its diagnostic and prognostic value, MCM7 may also serve as a therapeutic target. Since MCM7 plays a crucial role in DNA replication, targeting it could inhibit cancer cell proliferation, providing a novel approach to lung cancer treatment.[24] Samad et al. discussed the potential of targeting MCM7 in lung cancer therapy, noting that its inhibition could disrupt tumor growth by interfering with DNA replication processes.[25]
Cytological analysis of sputum increases the lung cancer detection rate, and hence, it is considered a poor man’s bronchoscopy.[26] Veena et al., in 2019, suggested an advanced technique for sputum processing to address the shortcomings of sputum cytology and the uncertainty of atypical samples.[27] Recently, there has been a lot of focus on finding certain molecular markers to add to the traditional morphological study of sputum to increase the technique’s sensitivity and specificity. LBC of sputum samples provide monolayered smear without much mucus and other debris in the smear background, which reduces the likelihood of diagnostic errors. Furthermore, the cells clogged in mucus can be obtained in the smear without much reactive changes. Computerized screening of sputum cytology slides may be possible with LBC. Cell blocks prepared in cell buttons processed in the LBC method provided clear sections without much background materials of mucus and inflammatory cells. In the current study, we have noticed a higher expression of MCM7 protein in all the lung cancer sputum samples analyzed, which was followed by MCM2 expression which supports the previous reports. MCM2 was found to be an independent predictor of survival in patients with NSCLC and a prospective diagnostic marker for premalignant lung lesions.[28,29] According to Toyokawa et al., both the messenger ribonucleic acid (mRNA) and protein levels of MCM7 expression were significantly greater in various cancer tissues compared to normal tissues.[10] However, we could not find MCM7 expression in benign respiratory epithelium in any of the sputum samples. The observation of higher MCM7 H score values being positively linked to tumor type in lung sputum samples is in consistence with other earlier reports in lung tissue samples from Liu et al.[30]
Our investigation also demonstrated a noteworthy correlation of MCM2 with tumor cytology and MCM 7 with tumor stage and cytological type. The prognosis can be predicted using this observation. These results corroborate earlier research that revealed MCM7 markers could be utilized to forecast tumor growth and patients’ prognosis with NSCLC.[10] A substantial association between MCM2 expression and a poor prognosis in NSCLC patients was also discovered by Yang et al., indicating that tumor proliferation may be a crucial factor in determining the prognosis of NSCLC.[31] The poor survival rate for NSCLC patients is associated with the late-stage presentation of patients with unresectable tumors because there is no validated screening method for early diagnosis, making small biopsies/cytology samples more essential.[32]
MCM protein sensitivity in sputum samples has also been examined as a potential predictor of malignant lesions. Cytology samples are more sensitive than histology samples because MCM-positive cells typically arise at the surface of the aberrant epithelia.[33] For this reason, MCMs are extremely promising indicators for the early identification of cancer and precancerous conditions in cytology samples. In our evaluation of sputum samples from patients with various subtypes of lung cancer, we found that the MCM7 protein demonstrated a sensitivity of 92% and a specificity of 100% for diagnosing malignancy. Another study showed that cell division cycle 6 protein demonstrated the highest sensitivity of 87.7% in sputum samples for characterization of malignant cells followed by MCM5 and MCM2 with 66.67% and 58.89%, respectively.[34]
The current study could recognize some of the samples of sputum reported with an indiscriminate diagnosis of atypical cells to a definite diagnosis of malignancy, suggesting the significance of MCM7 in the differential diagnosis of malignancy in sputum samples. This information can be used to define some alternative techniques for the detection of malignant cells in sputum, particularly for people with clinical complaints and abnormal X-ray results in low-resource environments. Incorporation of MCM7 immunocytochemistry in routine clinical practice will certainly enhance the diagnostic efficacy of sputum cytology, which is comparatively easier, cost-effective, and highly economical compared to bronchoscopy and biopsy for the diagnosis of lung cancer, and it can be repeated easily if required without any hardship to the patient. While this research offers insightful information about using MCM2 and MCM7 as biomarkers to enhance traditional sputum cytology, there are some limitations which include the relatively small sample size in each type of tumors, lack of early-stage tumors and precancerous conditions, and lack of follow-up. A multicentric study in a large cohort from a broader population with follow-up of patients is warranted to generalize the significance of MCM proteins in lung cancer diagnosis.
SUMMARYTo our knowledge, this is the first study to report MCM7 expression in sputum cells, highlighting its potential to significantly increase the sensitivity of sputum cytology. The incorporation of MCM7 protein analysis into conventional sputum cytology could greatly improve the detection of malignant cells, thus supplementing morphological evaluation methods. This is particularly valuable for samples with atypical cells, where traditional cytology alone might not provide definitive results. By increasing the sensitivity of sputum cytology, MCM7 can help ensure that more cases of lung cancer are accurately identified at an earlier stage. Furthermore, our analysis suggests that examining MCM7 expression in sputum samples from patients with biopsy-proven premalignant lung conditions could establish this protein as a useful marker for primary screening of lung cancer. This application has the potential to facilitate earlier detection and intervention, thereby improving patient outcomes. In addition to its diagnostic applications, the potential of MCM7 in forecasting the clinical behavior of lung lesions, as well as its prognostic value in terms of survival, warrants further investigation. Exploring these aspects could pave the way to the development of more precise and personalized treatment approaches for lung cancer patients, ultimately enhancing prognosis and survival rates.
AVAILABILITY OF DATA AND MATERIALSThe data analyzed in this study can be accessed through the corresponding author upon reasonable request.
ABBREVIATIONSAUC: Area under the curve
CDC6: Cell division cycle 6
CT: Computed tomography
DPX: Dibutylphthalate polystyrene xylene
Ki-67: Kiel-67
LBC: Liquid-based cytology
LUAD: Lung adenocarcinoma
LUSC: Lung squamous cell carcinoma
MCM2: Minichromosome maintenance protein 2
MCM7: Minichromosome maintenance protein 7
MCMs: Minichromosome maintenance proteins
NMC: Negative for malignant cells
NPV: Negative predictive value
NSCLC: Non small cell lung carcinoma
PCNA: Proliferating Cell Nuclear Antigen
PPV: Positive predictive value
ROC: Receiver operating characteristic
SCLC: Small cell lung carcinoma
AUTHOR CONTRIBUTIONSNP: Wet lab works, data collection and analysis, interpretation of results, original manuscript preparation, review and editing; AL: Supervision of wet lab works, discussion drafting; JKKM: Statistical data analysis; VVS: Immunocytochemical data analysis; KS: Conceptualization, supervision, resources, review, and editing. All authors read and approved the final manuscript.
Comments (0)