In 2008, a mysterious outbreak of a haemorrhagic fever virus disease occurred in Zambia and South Africa killing 80% of the infected people (1). The origin and host of the virus later identified as Lujo virus remains unknown to date. This and several other examples continuously indicate that infectious agents (zoonoses) originating from domestic animals and wildlife present profound threats to global public health and trade (2). These diseases, often unpredictable and challenging to treat, have been increasingly linked to wildlife (3). Since 1940, approximately 60% of human infectious diseases have emerged from animal reservoirs, with more expected to cross into human populations by 2070 or earlier (4–6). These zoonotic pathogens cause a billion of human illnesses and millions of deaths annually (4, 7), transmitted through various means including consumption of infected food (fruits, meats, vegetables, and date palm sap), aerosols, secretions (saliva), excreta (urine and faeces), and handling animals and their products (8–11). The magnitude and complexity of these transmission pathways underscore the urgent need for comprehensive surveillance and robust public health interventions to mitigate these escalating risks.
Over the past seven decades, certain animals have been identified as key carriers of zoonotic pathogens. Bats, rodents, and non-human primates (NHPs) are particularly significant due to their widespread presence and close interactions with humans (12–15). Bio-diversity loss related to agriculture, environmental changes, and other commercial activities have been linked to increased contact between humans and wildlife and cross-species transmission of pathogens (2–4). Bats and rodents, in particular, often live commensally or semi-commensally in human dwellings (8, 16). Bats, capable of long-distance flight, spread viruses such as Nipah (17) and Marburg (18), and are implicated in the origins of Ebola virus (19) and severe acute respiratory syndrome (SARS) coronaviruses −1 and 2 (SARS-CoV-1 and SARS-CoV-2) (20, 21). On the other hand, Lassa virus (LASV) and Yersinia pestis are among the deadly pathogens directly linked to rodents (22, 23) while Lujo virus (LUJV) is suspected to have a rodent reservoir (24). The unpredictable emergence of LUJV, a hemorrhagic fever virus first identified in 2008, underscores the critical importance of vigilance and review of information, as its host and transmission dynamics remain unknown. Meanwhile, NHPs, due to their genetic similarities to humans and increasing habitat overlap due to habitat losses linked to agriculture, are notably implicated in the zoonotic origin of HIV and may serve as potential intermediaries of Ebola virus, particularly during an outbreak (25–28). Addressing these complex interactions demands sustained monitoring and proactive public health strategies to mitigate the evolving risks posed by zoonotic pathogens.
Given the substantial evidence of zoonotic threats posed by these animals (12–15), this study reviewed literature from January 1990 to December 2022 to examine the epidemiology and public health implications of pathogens in bats, rodents, and NHPs in Zambia. In the light of these threats, weak health systems with little capacity for multi-pathogen laboratory diagnosis (29), aggregated data may help identify hotspots which may guide presumptive clinical diagnosis in specific regions, and enhance our ability to respond to outbreaks effectively. The organized information may also inform policymakers, public health practitioners, clinicians, researchers, and financial stakeholders, providing critical insights for diagnosis, treatment, future actions and research in order to safeguard human and animal health.
2 Methods 2.1 Information sources and search strategiesA comprehensive data search was conducted across three electronic databases including (PubMed, Google Scholar, and CiNii Articles Incorporated Database) to identify articles and accompanying data reporting pathogens in bats, rodents, and NHPs. The search was restricted to studies published between January 1990 and December 2022. The period was selected in order to provide a comprehensive and nuanced understanding of the research trends and distribution of pathogens in the light of continuous changes in globalisation, pandemics, research tools national priorities, data availability, and demographic changes. Eligible original studies were identified using the following search terms with the help of Boolean operators (AND, OR): ((Virus) AND (Zambia)) AND (bat), ((Virus) AND (Zambia)) AND (rodent), ((Virus) AND (Zambia)) AND (non-human primates), ((Virus) AND (Zambia)) AND (baboons), ((Virus) AND (Zambia)) AND (Monkeys), ((Bacteria) AND (Zambia)) AND (bats), ((Bacteria) AND (Zambia)) AND (rodent), ((Bacteria) AND (Zambia)) AND (non-human primates), ((Bacteria) AND (Zambia)) AND (baboons), ((Bacteria) AND (Zambia)) AND (Monkeys), ((Protozoa) AND (Zambia)) AND (bats), ((Protozoa) AND (Zambia)) AND (rodent), ((Protozoa) AND (Zambia)) AND (non-human primates), ((Protozoa) AND (Zambia)) AND (baboons), ((Protozoa) AND (Zambia)) AND (Monkeys), ((Zoonoses) AND (Zambia)) AND (bats), ((Zoonoses) AND (Zambia)) AND (rodent), ((Zoonoses) AND (Zambia)) AND (non-human primates), ((Zoonoses) AND (Zambia)) AND (baboons), ((Zoonoses) AND (Zambia)) AND (Monkeys), ((Pathogen) AND (Zambia)) AND (bats), ((Pathogen) AND (Zambia)) AND (rodent), ((Pathogen) AND (Zambia)) AND (non-human primates), ((Pathogen) AND (Zambia)) AND (baboons), and ((Pathogen) AND (Zambia)) AND (Monkeys). The search strategy also included ((bats OR rodents OR “non-human primates” OR baboons OR monkeys) AND (bacteria OR protozoa OR zoonoses) AND (Zambia) AND (prevalence OR seroprevalence OR wildlife OR national park)) Following identification of some articles which specified certain pathogens, search terms were extended to include some names of identified pathogens as follows: ((bats OR rodents OR “non-human primates” OR baboons OR monkeys) AND (bacteria OR protozoa OR zoonoses OR Trypanosomes OR Giardia OR Cryptosporidium OR Coxiella OR Borrelia OR Paramyxovirus OR Leptospira OR Filovirus OR “Marburg virus” OR “Hepatitis virus” OR Rotavirus OR Arenavirus OR Rickettsia OR Babesia OR Anaplasma) AND (Zambia)).
2.2 Inclusion criteriaSelected studies focused on rodents, bats, or NHPs in Zambia, reporting on sample size and type, diagnostic methods, and pathogen type. Studies had to be peer-reviewed, in English, fully accessible, conducted in Zambia, and published from January 1, 1990, to December 31, 2022. Experimental studies that combined both surveillance and experimentation were also included.
2.3 Exclusion criteriaStudies were excluded if they lacked detailed methodological descriptions, did not report prevalence data, were not peer-reviewed, or were not written in English. Journal articles published outside the predetermined review period, and duplicates as well as non-original research were excluded from the review.
2.4 Data extraction and managementData were independently extracted for each pathogen or potential pathogen by two reviewers (SMM and BM) using a standardised form for this review. It included animal species (rodent, bat, NHP), sample size, type, diagnostic methods, pathogen type, positives, and genetic matches via BLAST. Other data covered study location, design, publication year, authors, and journal. Discrepancies were resolved through discussion or a third reviewer who was available. Data were managed in Microsoft Office Excel 2018, ensuring integrity with backups. Zotero (Version 5.0.96.3) stored study details including titles, abstracts, authors, years, journals, and extracted PDFs where available, facilitating comprehensive information retrieval and management. The quality of the studies was evaluated using the JBI’s critical appraisal tools for prevalence and incidence studies to assess the trustworthiness, relevance, and results of the studies (30).
2.5 Data synthesis and analysisThe prevalence rate of pathogens in selected animal species was directly extracted from the reviewed articles. For studies reporting data on multiple animal species, the prevalence was recalculated separately for each species of interest to ensure accurate representation (31–33). In instances where data from multiple studies were available, the cumulative prevalence of certain pathogens was determined by combining data from these studies. The public health risk associated with each reported pathogen was assessed based on existing evidence from the included articles and relevant literature demonstrating the pathogen’s potential to cause disease.
2.6 Data presentationDescriptive data were summarised in tables showing the number of samples analysed, number of positive cases, and prevalence rates. Maps were created to show the distribution of sample collection sites across Zambia and reported highly infectious agents. A detailed summary of findings was presented, including a discussion and implications for future research.
2.7 Ethical considerationsAll included studies were evaluated for adherence to ethical standards for animal research. This included reviewing whether the original studies obtained appropriate ethical approvals and consent for animal use.
3 Results 3.1 Literature searchThe literature search identified 37 eligible articles. The number represents the actual number of surveillance studies conducted specifically on bats, rodents, and NHPs in Zambia from 1990 to 2022.
3.2 Publication trendsA total of thirty-seven original research articles were included in the analysis, covering the period January 1, 1990 to December 31, 2022. The search did not yield any articles within the investigated databases for the period 1990 to 2009. The highest publication frequency was observed in the years 2018 and 2019 (n = 10). When categorised by host type, 41%% (14/39) of the articles represented studies focused on bats (34–47), 30.8% (12/39) on rodents (23, 31, 48–56), and 25.6% (10/39) on NHPs (32, 57–65). One article (2.6%; 1/39) reported on both NHPs and rodents (33). In terms of pathogens detected, 66.7% (26/39) of the articles investigated viruses exclusively (31–34, 37–40, 43–47, 50–52, 55, 58–64), 17.9% (7/39) focused solely on bacteria (23, 35, 36, 41, 48, 53, 54), and 5.1% (2/39) protozoa alone (42). The remaining 10.3% (4/39) of the articles addressed mixed infections involving bacteria and viruses (49), helminths and viruses (56), as well as bacteria and protozoa (57).
3.3 Study sites, sampling approaches, and sample typesSamples were collected from animals in study sites across 14 out of the 116 districts in Zambia (Figure 1). Reasons for selecting the reported study sites were not indicated. All the articles reported use of cross-sectional study designs. Short-term cross-sectional studies accounted for 94.9% (37/39) of all reported articles compared to 5.4% (2/37) of long term cross-sectional studies (38, 59). Almost all articles (97.4%, 38/39) had spleen, liver, kidneys, and blood as samples of choice with the exception of one study (2.6%, 1/39) in rodents that extended its investigations to semen (56).

Figure 1. Sampling sites, derived from reviewed articles, for pathogens detected in bats, rodents, and NHPs across Zambia. Districts in which only rodents were sampled (Blue), bats only (Brown), rodents and NHPs (lime green), rodents and bats (red) and all animal types (Green).
3.4 Diversity and geographical distribution of reported bat, rodent, and NHP speciesThirteen species of bats were reported within the study areas in Livingstone, Lusaka, Monze, Ndola and Serenje districts (34–47). The reported species of bats included Hipposideros gigas, Hipposideros vittatus, Miniopterus schreibersii, Rousettus aegyptiacus, Minipteros sp., Myotis sp., Rhinolophus simulator, Macronycteris vittatus, and other unknown species in the genera Rhinolophus and Hipposideros in Lusaka province where most of the investigations took place; Eidolon helvum in Ndola and Serenje (Kasanka National Park) districts; Epomophorus crypturus in Monze district; and Nycteris sp. in Livingstone district (34–47). In terms of common species, Rousettus aegyptiacus (23.2%, 719/3100) and Eidolon helvum (68.2%, 2114/3100) were the most captured species of bats, accounting for 91.4% (2,833/3100) of all reported bats.
Rodents belonging to 17 genera were identified in sampling sites in Chibombo, Kafue, Livingstone, Lusaka, Mazabuka, Mfuwe, Mpulungu, Namwala, Nyimba, Serenje, Sinda, and Solwezi districts. Reported rodent species included Acomys subspinosus, Aethomys chrysophilus, Arvicanthis sp., Cricetomys gambianus, Gerbilliscus leukogaster, Grammomys sp., Graphiurus sp., Hylomyscus alleni., Praomys sp., Lemniscomys rosalia, Mastomys natalensis, Mus minutoides, Otomys sp., Rattus rattus, Saccostomus campestris, Pelomys sp. (23, 31, 33, 48–53, 55). The distribution of rodents was fairly ubiquitous across the districts except for Mus minutoides which was only reported in Lusaka district. The most trapped rodent species was M. natalensis which accounted for 91.4% (1825/1996) of all captured rodents.
Chlorocebus and Papio were the only genera of NHPs reported in the reviewed articles (32, 57, 59–61, 64). The genus Chlorocebus was represented by Chlorocebus pygerythrus (vervet monkey) and Chlorocebus cynosures. Meanwhile, kinda yellow baboon (Papio kindae), yellow baboons (Papio cynocephalus), and Chacma baboons (Papio ursinus) comprised NHPs in the genus Papio. All species were reported in the sampling sites in Livingstone and Mfuwe districts except in the Kafue National Park where only Chlorocebus cynosures were sampled. The most captured NHP was P. cynocephalus (28.3%, 339/1198).
3.5 Diversity of infectious agents reported in bats, rodents, and NHPsA combined total of 50 distinct infectious agents were reported in the reviewed articles. Bats and rodents each accounted for 36% (18/50) of all infectious agents whereas NHPs were responsible for 28% (14/50). Viruses were primarily the most frequently reported types of microorganisms accounting for 62% (31/50), mirroring the focus of the research in the reviewed articles. A total of 18% (9/50) of infectious agents were bacteria and the remaining 20% (10/50) were parasites. The array of infectious agents across bats, rodents, and NHPs represented highly infectious pathogens (Figure 2) and those with unknown zoonotic potential (Tables 1–4; Supplementary Table S1).
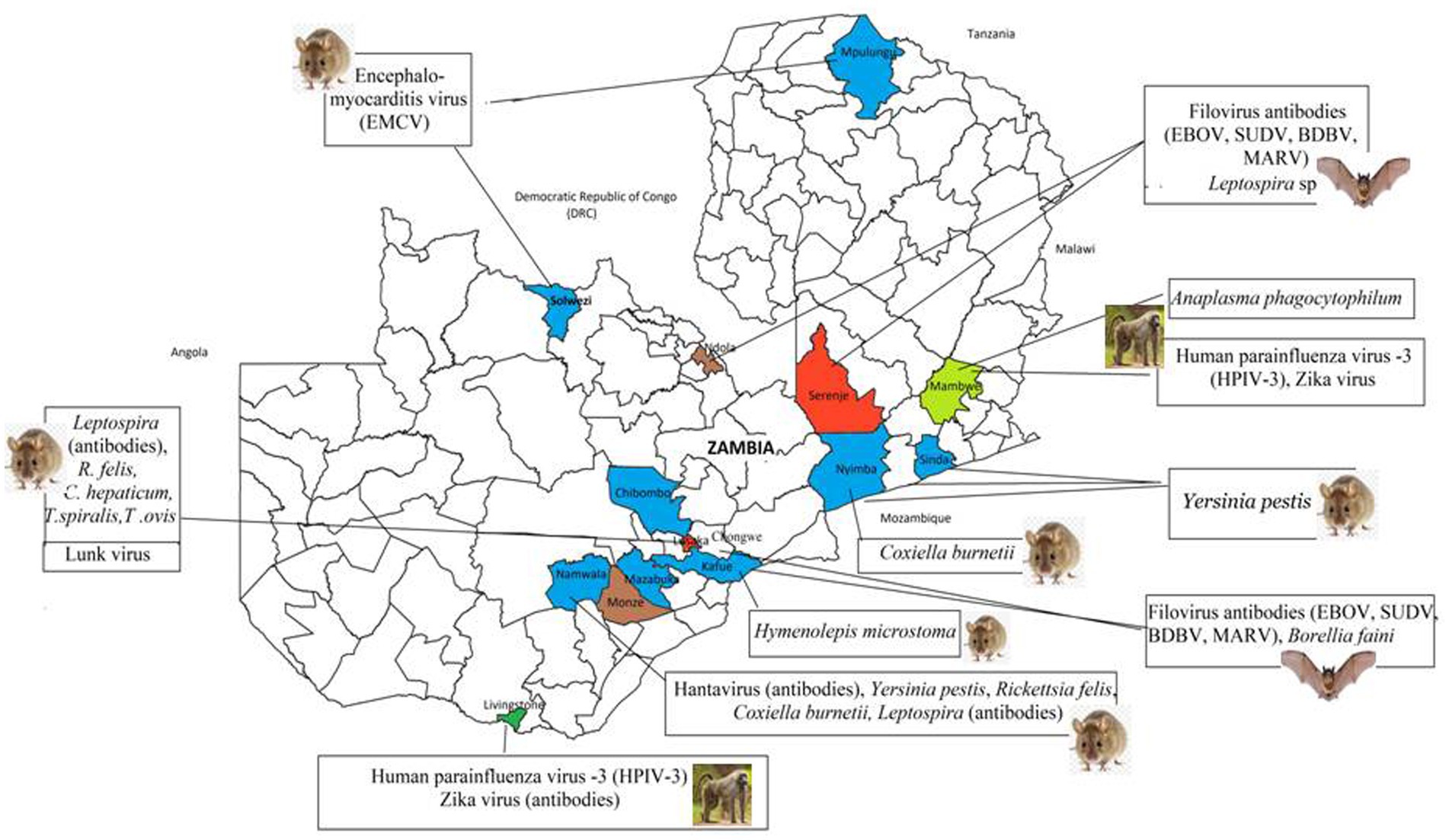
Figure 2. Distribution of high risk infectious agents reported in wildlife in the reviewed articles. Coloured areas represent the location of study sites from the 37 articles.
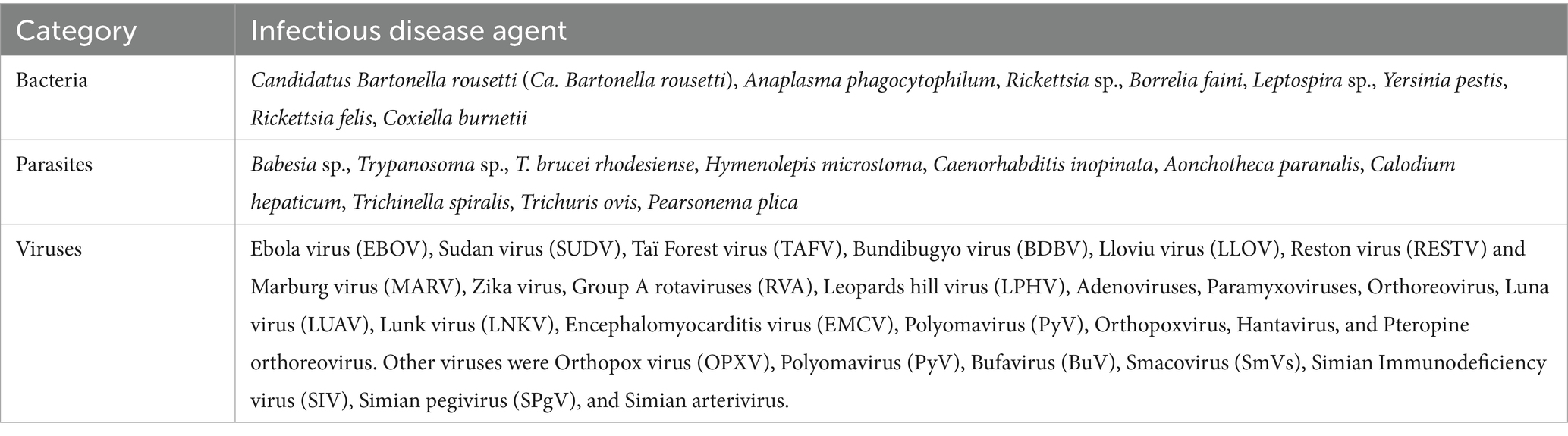
Table 1. Diversity of bacteria, parasites, and viruses reported in the reviewed articles.
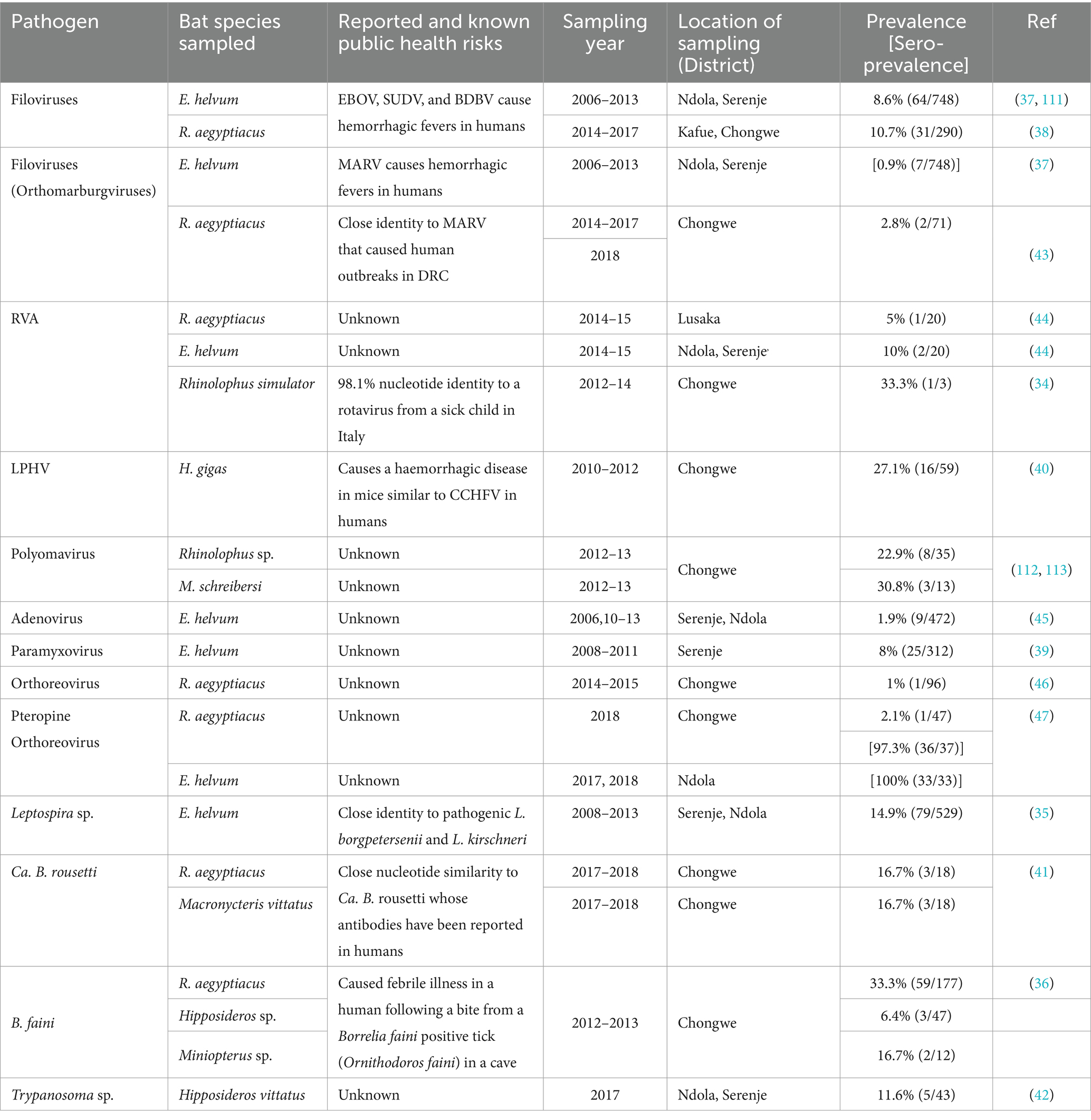
Table 2. Viral, bacterial, and protozoan pathogens reported in bats in Zambia, 1990 to 2022.
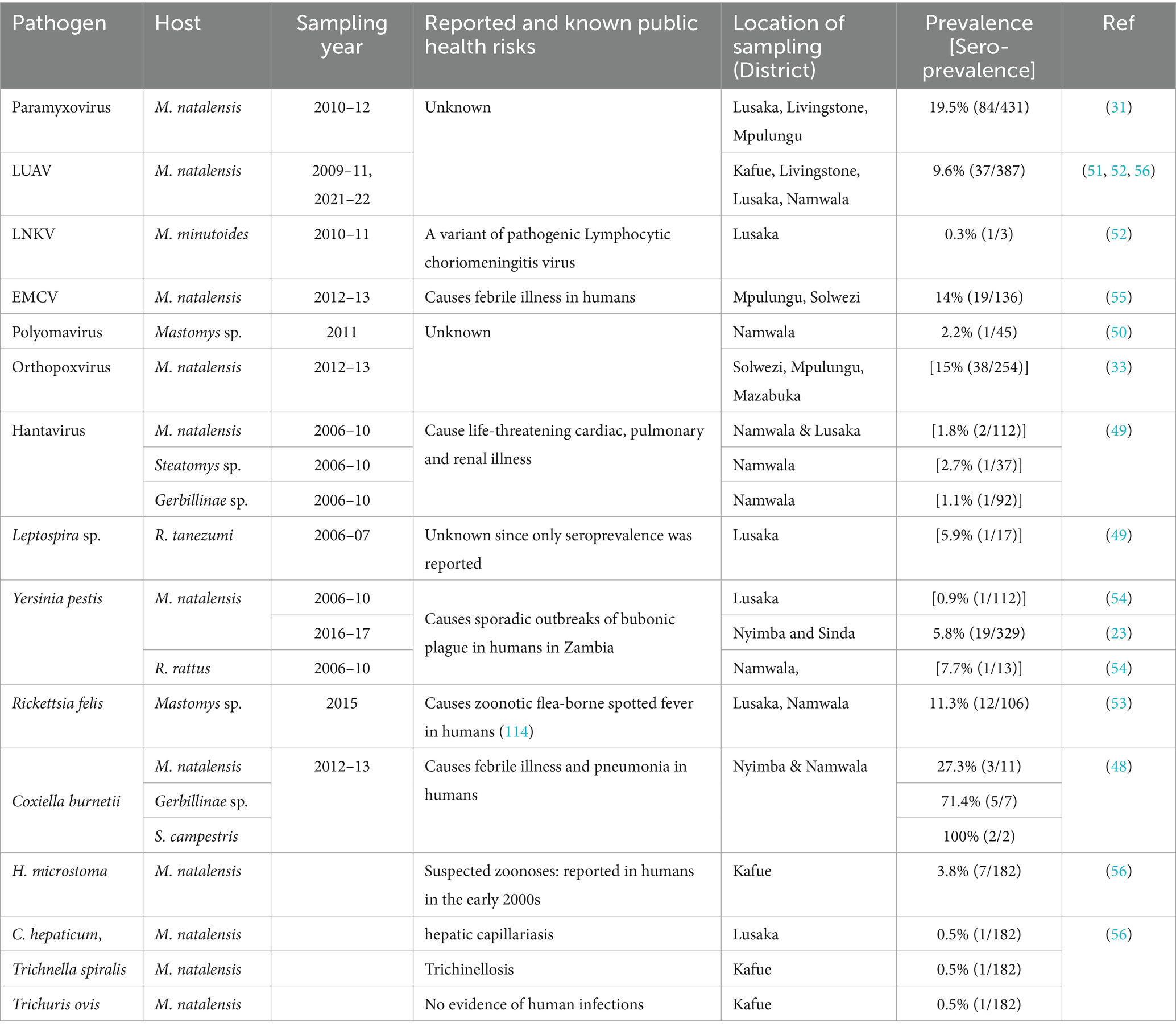
Table 3. Viral and bacterial pathogens reported in rodents in Zambia, 1990 to 2022.

Table 4. Viral, bacterial, and protozoan pathogens reported in NHPs in Zambia, 1990 to 2022.
3.6 Distribution and zoonotic potential of reported infectious agents by animal typeThe 18 infectious agents reported in bats (Table 1) were primarily in Rousettus aegyptiacus and Eidolon helvum, comprising 77.8% (14/18) viruses, 16.7% (3/18) bacteria, and 5.5% (1/18) parasites. Noteworthy zoonotic viruses included EBOV, MARV, RVA, Borrelia faini, Leptospira sp., among others. In rodents, viruses and parasites each contributed 38.9% (7/18) to the reported infectious agents compared to 22.2% (4/18) by bacteria (Table 2). Most pathogens were reported in Mastomys natalensis. Notable pathogens were Yersinia pestis, Rickettsia felis, Coxiella burnetti, Calodium hepaticum, and Leptospira sp. Among NHPs, 71.4% (10/14) of the reported infectious agents were viruses (Table 3). Bacteria and parasites each accounted for 14.3% (2/14). Human parainfluenza virus type 3 (HPIV3), filoviruses, Trypanosoma brucei rhodesiens, and Anaplasma phagocytophilum were the headline pathogens.
4 DiscussionThis review uncovered genetic and serological evidence of a worrying array of infectious agents, some with documented history of causing severe human illnesses in Zambia (23, 36, 66) and elsewhere (67–69). Among these were EBOV, SUDV, BDBV, MARV, RVA, ZIKV, HPIV3, EMCV, Hantavirus, Leptospira sp., Ca. B. rousetti, B. faini, Y. pestis, R. felis, C. burnetti, A. phagocytophilum, and T. b. rhodesiense (65, 67, 70–83). Three pathogens, Y. pestis (84), B. faini (36), and T. b. rhodesiense were detected in active human clinical illness alongside their presence in wildlife, highlighting the immediate threat these pathogens pose. Furthermore, LPHV and LNKV (40, 52) were flagged as public health concerns due their similarity in clinical symptoms observed in humans and animals (85), and their close genetic relationships with known highly pathogenic organisms (86), respectively. These findings emphasise the critical need for enhanced surveillance and comprehensive public health strategies to prevent and control potential outbreaks.
4.1 Quality of reviewed articles and research trendsAll the articles included in the review utilized standard polymerase chain reaction (PCR) detection methods and reported the prevalence of pathogens, except for one study that did not (63). Sampling of non-human primates (NHPs) was predominantly focused on national parks in the Livingstone and Mfuwe districts, where the researchers had active projects. Additionally, the studies exhibited a bias toward viral pathogens, reflecting the specific interests of the researchers. Articles detailing zoonotic pathogen circulation in the animals of interest from 1990 to 2009 were absent. Although the reason for the absence of articles during that period could be linked to various factors, data from some review articles suggest a possible focus on anthrax, bovine tuberculosis, trypanosomiasis, tick-borne parasitic diseases, and other zoonoses in livestock (87–91). Therefore, there is need for further investigations over the observed trends in order to fully understand the evolution of research on zoonoses in Zambia. Nonetheless, post-2009, there was a notable surge in research involving bats, rodents, and NHPs (31–33, 35, 37, 39, 40, 49–52, 57, 60, 62). This increase in research activity likely stems from significant events such as the emergence of LUJV in Zambia in 2008 (1), the global swine influenza outbreak in 2009 (92), and the recurring, deadly outbreaks of Marburg virus disease (MDV) (70), and EVD in the neighbouring DRC (82).
4.2 Sampling approaches in the reviewed articlesCross-sectional study designs were the most predominant, although they lacked subsequent follow-up research, except for two studies (37, 38). Therefore, the temporal dynamics related to abundance and likelihood of outbreaks of most reported pathogens remain poorly studied. These glaring gaps emphasise the critical need for robust long term surveillance studies in order to fully understand the spatial, temporal, and transmission dynamics of infectious agents within their micro-environments (56).
4.3 Organ-specific distribution of pathogensUnderstanding organ-specific pathogen distribution enhances targeted surveillance and diagnostic strategies, crucial for predicting and controlling infectious diseases. Most studies focused on spleen, liver, kidneys, and blood, with one exception using semen samples (56), highlighting their potential for pathogen detection, including viruses like EBOV and LASV (56, 93–95). This underscores the need to screen multiple organs and fluids to increase detection rates. For filovirus surveillance in bats, semen and seminal vesicles may offer valuable insights, as traditional samples (blood, liver, etc.) have scarcely yielded positive RNA results (14, 19). Integrating diverse sample types could enhance disease surveillance and response efforts significantly.
4.4 Geographical distribution of reported pathogensThe distribution of pathogens mirrored the spread of their hosts. However, reservoir presence did not always correlate with infectious agents (51, 52, 56). Studies covered just 14 of 116 districts, notably neglecting Western and Luapula provinces with rich river systems ideal for diverse pathogens (56). This gap raises concerns about potential undetected zoonotic hotspots. Sparse surveillance hampered understanding of pathogen spread, crucial for guiding presumptive treatment in areas lacking multi-pathogen diagnostic capabilities in healthcare settings.
4.5 Public health concerns of reported viral pathogensSerological and molecular evidence of bats and NHPs encountering pathogenic filoviruses raises significant public health concerns. Serological findings are notable as EBOV RNA has only been detected once in bats (19). EBOV and MARV cause deadly, unpredictable epidemics in Central and West Africa, with the 2014–2015 West African EBOV outbreak causing over 11,310 deaths (73, 96). In Zambia, spill-over risk stems from R. aegyptiacus bats, and migratory E. helvum bats from filovirus-endemic Congo Basin countries (97). Infected humans crossing the open border from the EBOV hotspot, DRC, are potential sources of human to human transmission (82). Wild RVAs (34, 44) also raise public health worries due to their potential impact on current vaccines (98), particularly the possible emergence of reassorted viruses not covered by existing vaccines (99). The discovery of an RVA with 98.1% nucleotide similarity to shows the potential cross species transmission between bats and humans probably through contaminated shared fruits and water (34). On the other hand, HPIV3, Hantavirus, Zika virus, and novel pathogens such as LPHV and LNKV cannot be ignored. Discovery of an HPIV3 sequence in baboons, closely resembling a strain isolated from a sick child in Saudi Arabia (60, 100), underscores the need for investigations into zoonotic HPIV3 in children in Zambia. Therefore, both known and novel viruses reported herein, demand rigorous investigation to mitigate their potential threat to local and global health.
4.6 Public health concerns of reported bacterial pathogensYersinia pestis, C. burnetii, A. phagocytophilum, and Leptospira sp. were the headline pathogens in the reviewed articles. Yersinia pestis poses a documented threat in Zambia, causing sporadic outbreaks of bubonic plague, particularly in eastern regions (23, 54). On the other hand, C. burnetii, responsible for Q fever which has never been reported in humans in Zambia, seems widespread within multiple rodent hosts in diverse ecological settings (101). Leptospira sp., similar to other zoonotic pathogens, suffers from a significant paucity of molecular and human data in Zambia. This calls for public health vigilance due to recent outbreaks in humans in neighbouring Tanzania (102). Anaplasma phagocytophilum was another significant zoonotic pathogen reported in wildlife. It is a multi-host intracellular pathogen, which causes varying severity of human febrile granulocytic anaplasmosis and encephalitis (103, 104). Like the emergence of B. faini which caused Zambia’s first clinical case of human borreliosis (36), its detection is alarming and warrants further investigations. The disease caused by B. faini presented with symptoms resembling malaria and flu, including high fever, initially confusing diagnosis at a local clinic, a common phenomenon with zoonotic pathogens (105). B. faini formed a monophyletic lineage akin to relapsing fever borreliae in the USA (36). Further investigations are critically needed to understand its overall impact on human health in Zambia.
4.7 Public health concerns of reported parasitic pathogensSignificant public health threats from pathogens including T. b. rhodesiense, C. hepaticum, T. spiralis, and T. ovis still exist as reported in the articles. Human African trypanosomiasis (HAT) caused by T. b. rhodesiense provided a problematic diagnostic process among clinicians, resulting in delayed treatment, suggesting need for refresher courses on this neglected zoonoses (65, 66). Strategic awareness campaigns may be helpful to educate the public about HAT and pathogenic parasites such as C. hepaticum, T. spiralis, and T. ovis (106–110).
4.8 Implication for the Southern Africa region and beyondIn the context of Zambia’s open boarders and its role as a key transportation hub for Southern Africa, the implications of the existence of highly pathogenic zoonotic organisms with potential for major outbreaks are even alarming. It significantly heightens the risk of zoonotic pathogen outbreaks spreading rapidly across the region. The free international connectivity and high traffic can swiftly transform a local outbreak into a pandemic. Economically, the region could suffer from costly public health crises, disrupting trade and travel, and impacting regional stability similar to COVID-19 (1). To mitigate the risk of zoonotic disease spread through these transportation hubs, robust screening and surveillance systems are vital. Continuous and consistent aggregation and analysis of available data on circulating zoonotic pathogens can help map hotspots and enhance the country and region’s ability to respond effectively.
5 Conclusion and future perspectivesThe findings underscore Zambia’s critical public health challenge: diverse pathogens with zoonotic potential identified in bats, rodents, and non-human primates across a few districts. With only 14 out of 116 districts reporting data, much of the country’s wildlife pathogen landscape remains unexplored. Studies from which the data was extracted were characterised by short-term investigations lacking follow-up investigations and detailed analysis of the micro-characteristics of host habitats. This gap leaves the ecology of most reported pathogens poorly understood. To address this, future studies should comprehensively examine all aspects of pathogen prevalence, transmission, and persistence dynamics to identify potential hotspots, both spatially and temporally. Additionally, there is a huge gulf between what is known about zoonotic pathogens in wildlife and in humans in Zambia. Thus, comprehensive serological and molecular studies are urgently needed to reveal the true burden of these pathogens on human health to guide laboratory diagnosis and region-specific treatment options.
Data availability statementThe original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributionsSaM: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. BM: Formal analysis, Funding acquisition, Resources, Validation, Writing – review & editing. KC: Formal analysis, Validation, Visualization, Writing – review & editing. JT: Formal analysis, Validation, Writing – review & editing. RH: Formal analysis, Validation, Writing – review & editing. MB: Formal analysis, Validation, Writing – review & editing. SC: Formal analysis, Methodology, Validation, Writing – review & editing. SoM: Formal analysis, Validation, Writing – review & editing. ES: Conceptualization, Formal analysis, Methodology, Supervision, Validation, Writing – review & editing.
FundingThe author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was partially supported by a grant (Grant number 4500474987) from the World Academy of Sciences for the advancement of science in developing countries awarded to the co-author, Benjamin Mubemba. The funders had no role in the publication process nor choice of the journal.
Conflict of interestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s noteAll claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materialThe Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2024.1471452/full#supplementary-material
References1. Simulundu, E, Mweene, AS, Changula, K, Monze, M, Chizema, E, Mwaba, P, et al. Lujo viral hemorrhagic fever: considering diagnostic capacity and preparedness in the wake of recent Ebola and Zika virus outbreaks. Rev Med Virol. (2016) 26:446–54. doi: 10.1002/rmv.1903
PubMed Abstract | Crossref Full Text | Google Scholar
2. Allen, T, Murray, KA, Zambrana-Torrelio, C, Morse, SS, Rondinini, C, Di Marco, M, et al. Global hotspots and correlates of emerging zoonotic diseases. Nat Commun. (2017) 8:1124. doi: 10.1038/s41467-017-00923-8
PubMed Abstract | Crossref Full Text | Google Scholar
3. Allocati, N, Petrucci, AG, Di Giovanni, P, Masulli, M, Di Ilio, C, and De Laurenzi, V. Bat–man disease transmission: zoonotic pathogens from wildlife reservoirs to human populations. Cell Death Discov. (2016) 2:16048. doi: 10.1038/cddiscovery.2016.48
PubMed Abstract | Crossref Full Text | Google Scholar
4. Delia, G, Mutua, F, Ochungo, P, Kruska, R, Jones, K, Brierley, L, et al. Mapping of poverty and likely zoonoses hotspots International Livestock Research Institute – Zoonoses Project. Nairobi, Kenya. (2012).
5. Carlson, CJ, Zipfel, CM, Garnier, R, and Bansal, S. Global estimates of mammalian viral diversity accounting for host sharing. Nat Ecol Evol. (2019) 3:1070–5. doi: 10.1038/s41559-019-0910-6
Crossref Full Text | Google Scholar
6. Carlson, CJ, Albery, GF, Merow, C, Trisos, CH, Zipfel, CM, Eskew, EA, et al. Climate change increases cross-species viral transmission risk. Nature. (2022) 607:555–62. doi: 10.1038/s41586-022-04788-w
PubMed Abstract | Crossref Full Text | Google Scholar
7. Smith, KM, Machalaba, CC, Seifman, R, Feferholtz, Y, and Karesh, WB. Infectious disease and economics: the case for considering multi-sectoral impacts. One Health. (2019) 7:100080. doi: 10.1016/j.onehlt.2018.100080
PubMed Abstract | Crossref Full Text | Google Scholar
8. Fichet-Calvet, E, Lecompte, E, Koivogui, L, Soropogui, B, Doré, A, Kourouma, F, et al. Fluctuation of abundance and Lassa virus prevalence in Mastomys natalensis in Guinea, West Africa. Vector Borne Zoonotic Dis. (2007) 7:119–28. doi: 10.1089/vbz.2006.0520
PubMed Abstract | Crossref Full Text | Google Scholar
9. Luby, SP, Rahman, M, Hossain, MJ, Blum, LS, Husain, MM, Gurley, E, et al. Foodborne transmission of Nipah virus, Bangladesh. Emerg Infect Dis. (2006) 12:1888–94. doi: 10.3201/eid1212.060732
PubMed Abstract | Crossref Full Text | Google Scholar
10. Subramanian, M. Zoonotic disease risk and the Bushmeat trade: assessing awareness among hunters and traders in Sierra Leone. EcoHealth. (2012) 9:471–82. doi: 10.1007/s10393-012-0807-1
PubMed Abstract | Crossref Full Text | Google Scholar
11. Islam, MS, Sazzad, HMS, Satter, SM, Sultana, S, Hossain, MJ, Hasan, M, et al. Nipah virus transmission from bats to humans associated with drinking traditional liquor made from date palm sap, Bangladesh, 2011-2014. Emerg Infect Dis. (2016) 22:664–70. doi: 10.3201/eid2204.151747
PubMed Abstract | Crossref Full Text | Google Scholar
12. Dahmana, H, Granjon, L, Diagne, C, Davoust, B, Fenollar, F, and Mediannikov, O. Rodents as hosts of pathogens and related zoonotic disease risk. Pathogens. (2020) 9:202. doi: 10.3390/pathogens9030202
Crossref Full Text | Google Scholar
13. Dietrich, M, Mühldorfer, K, Tortosa, P, and Markotter, W. Leptospira and bats: story of an emerging friendship. PLoS Pathog. (2015) 11:e1005176–6. doi: 10.1371/journal.ppat.1005176
PubMed Abstract | Crossref Full Text | Google Scholar
14. Koch, LK, Cunze, S, Kochmann, J, and Klimpel, S. Bats as putative Zaire ebolavirus reservoir hosts and their habitat suitability in Africa. Sci Rep. (2020) 10:14268. doi: 10.1038/s41598-020-71226-0
PubMed Abstract | Crossref Full Text | Google Scholar
15. Robertson, SN, Cameron, AI, Morales, PR, and Burnside, WM. West Nile virus Seroprevalence in an outdoor nonhuman primate breeding Colony in South Florida. J Am Assoc Lab Anim Sci. (2021) 60:168–75. doi: 10.30802/AALAS-JAALAS-20-000029
PubMed Abstract | Crossref Full Text | Google Scholar
16. Ayivor, JS, Ohemeng, F, Tweneboah Lawson, E, Waldman, L, Leach, M, and Ntiamoa-Baidu, Y. Living with bats: the case of Ve Golokuati township in the Volta region of Ghana. J Environ Public Health. (2017) 2017:5938934–11. doi: 10.1155/2017/5938934
PubMed Abstract | Crossref Full Text | Google Scholar
17. Epstein, JH, Anthony, SJ, Islam, A, Kilpatrick, AM, Ali Khan, S, Balkey, MD, et al. Nipah virus dynamics in bats and implications for spillover to humans. Proc Natl Acad Sci USA. (2020) 117:29190–201. doi: 10.1073/pnas.2000429117
PubMed Abstract | Crossref Full Text | Google Scholar
18. Amman, BR, Bird, BH, Bakarr, IA, Bangura, J, Schuh, AJ, Johnny, J, et al. Isolation of Angola-like Marburg virus from Egyptian rousette bats from West Africa. Nat Commun. (2020) 11:510. doi: 10.1038/s41467-020-14327-8
PubMed Abstract | Crossref Full Text | Google Scholar
19. Leroy, EM, Kumulungui, B, Pourrut, X, Rouquet, P, Hassanin, A, Yaba, P, et al. Fruit bats as reservoirs of Ebola virus. Nature. (2005) 438:575–6. doi: 10.1038/438575a
PubMed Abstract | Crossref Full Text | Google Scholar
20. Delaune, D, Hul, V, Karlsson, EA, Hassanin, A, Ou, TP, Baidaliuk, A, et al. A novel SARS-CoV-2 related coronavirus in bats from Cambodia. Nat Commun. (2021) 12:6563. doi: 10.1038/s41467-021-26809-4
PubMed Abstract | Crossref Full Text | Google Scholar
22. Akhuemokhan, OC, Ewah-Odiase, RO, Akpede, N, Ehimuan, J, Adomeh, DI, Odia, I, et al. Prevalence of Lassa virus disease (LVD) in Nigerian children with fever or fever and convulsions in an endemic area. PLoS Negl Trop Dis. (2017) 11:e0005711. doi: 10.1371/journal.pntd.0005711
PubMed Abstract | Crossref Full Text | Google Scholar
23. Nyirenda, SS, Hang’ombe, BM, Simulundu, E, Mulenga, E, Moonga, L, Machang’u, RS, et al. Molecular epidemiological investigations of plague in Eastern Province of Zambia. BMC Microbiol. (2018) 18:2. doi: 10.1186/s12866-017-1146-8
PubMed Abstract | Crossref Full Text | Google Scholar
25. Gao, F, Bailes, E, Robertson, DL, Chen, Y, Rodenburg, CM, Michael, SF, et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. (1999) 397:436–41. doi: 10.1038/17130
Crossref Full Text | Google Scholar
26. Khabbaz, RF, Heneine, W, George, JR, Parekh, B, Rowe, T, Woods, T, et al. Infection of a laboratory worker with simian immunodeficiency virus. N Engl J Med. (1994) 330:172–7. doi: 10.1056/NEJM199401203300304
PubMed Abstract | Crossref Full Text | Google Scholar
27. Davies, TJ, and Pedersen, AB. Phylogeny and geography predict pathogen community similari
Comments (0)